PLAID: Multimodal protein generation via latent diffusion
Sources: http://bair.berkeley.edu/blog/2025/04/08/plaid, http://bair.berkeley.edu/blog/2025/04/08/plaid/, BAIR Blog
Overview
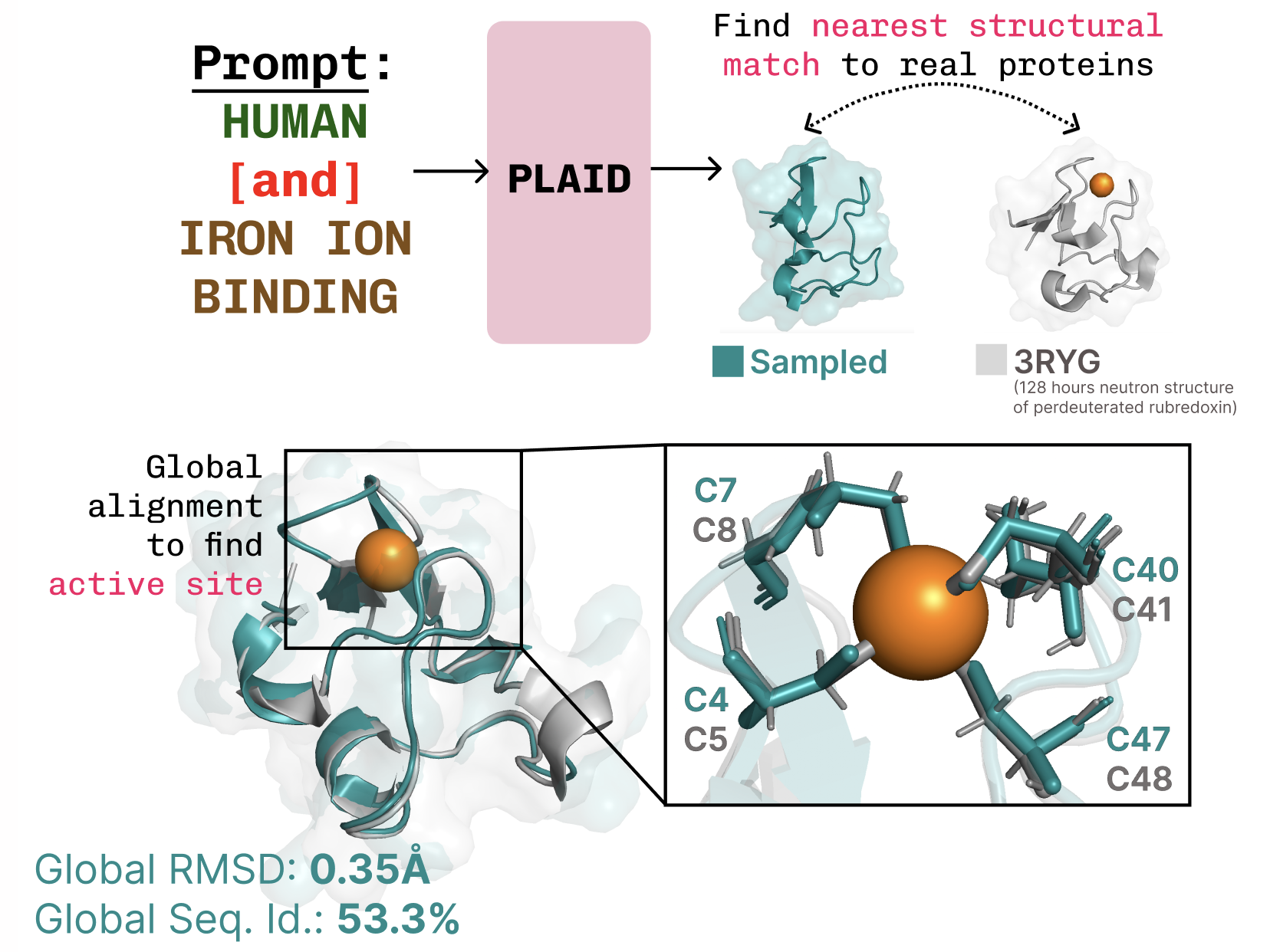
PLAID is a multimodal generative model that simultaneously generates protein 1D sequence and 3D structure by learning the latent space of protein folding models. The work follows the recognition of AI’s growing role in biology, exemplified by AlphaFold2’s Nobel Prize context, and asks what comes next after folding. PLAID learns to sample from the latent space of protein folding models to generate new proteins, and it can respond to compositional prompts that specify function and organism. Importantly, training can be performed on sequence data alone, since sequences are vastly more abundant than experimentally characterized structures. The core idea is to perform diffusion in the latent space of a folding model rather than directly in sequence or structure space. After sampling from this latent space of valid proteins, the model decodes both sequence and structure using frozen weights from a protein folding model. In practice, PLAID uses ESMFold—a successor to AlphaFold2 that replaces a retrieval step with a protein language model—as the structural decoder during inference. This setup leverages priors learned by a pretrained folding model to support protein design tasks while only requiring sequences during training. A notable aspect of the approach is addressing the high dimensionality of latent spaces in transformer models. The authors propose CHEAP (Compressed Hourglass Embedding Adaptations of Proteins) to learn a compression model for the joint embedding of protein sequence and structure. They find the latent space to be highly compressible and perform mechanistic interpretability to understand base-model knowledge, enabling all-atom generation in certain cases. While the work centers on sequence-to-structure generation, the authors emphasize potential extensions to multimodal generation for other domains with a predictor from a richer to a poorer modality. PLAID also offers an interface for control via prompts, inspired by how image generation can be guided with textual constraints. Although the ultimate aim is full textual control, the current system supports two axes of compositional constraints: function and organism. The authors demonstrate capabilities such as learning coordination patterns typical of metalloproteins (e.g., tetrahedral cysteine-Fe2+/Fe3+ motifs) while preserving sequence-level diversity. They compare PLAID samples with all-atom baselines and report better diversity and the capture of beta-strand patterns that are hard for some baselines to learn. If you’re interested in collaboration or wet-lab testing, the authors invite outreach. They also point to preprints and codebases for PLAID and CHEAP as resources for replication or extension.
Key features
- Multimodal generation: co-create protein sequences and all-atom structural coordinates
- Latent-diffusion workflow: sample from the latent space of a protein folding model, then decode
- Training on sequences only: leverage large sequence databases (2–4 orders of magnitude larger than structural data)
- Frozen-weights decoding: structure is decoded from sampled embeddings using frozen ESMFold weights
- CHEAP compression: learn a joint embedding compression for sequence–structure representations
- Function and organism prompts: compositional constraints guide generation
- Mechanistic interpretability: exploration of latent channels and emergent motifs (e.g., metal coordination patterns)
- Diversity emphasis: PLAID samples exhibit higher diversity and better motif recapitulation than some baselines
- Extensibility: the approach could be adapted to multimodal generation beyond proteins when an appropriate predictor exists
- Easy integration with existing priors: leverages priors from pretrained folding models similar to vision-language–action paradigms in robotics
Common use cases
- Design proteins with specified function and organism constraints (e.g., metalloproteins with targeted active-site motifs)
- Explore multimodal design spaces by prompting for function or species to bias sampling
- Generate diverse sequence–structure candidates that satisfy a function-structure relationship learned by the latent space
- Extend multimodal generation to more complex systems, such as protein complexes or interactions with nucleic acids or ligands, leveraging advances like AlphaFold3‑level capabilities
- Leverage the large sequence databases to train models that generalize to structure generation via latent representations rather than direct structure data
Setup & installation
Not provided in the source.
# Setup and installation information not provided in the source.Quick start
Note: This is a schematic illustration based on PLAID principles; exact APIs and runtimes will depend on the released codebases.
# PLAID quick-start (illustrative)
# This example shows the intended workflow rather than a drop-in, runnable script.
# 1) Sample a latent embedding from the diffusion model
z = sample_latent_embedding()
# 2) Decode sequence and structure using frozen ESMFold weights
sequence = decode_sequence_from_embedding(z)
structure = decode_structure_from_embedding(z, frozen_weights="ESMFold")
print(sequence)
print(structure)Pros and cons
- Pros
- Enables true co-generation of sequence and structure, guided by functional and organism prompts
- Leverages large-scale sequence data for training, mitigating the scarcity of structural data
- Uses frozen protein folding model weights for decoding, reducing the need to retrain complex structure modules
- CHEAP addresses the challenge of compressing a high-dimensional joint embedding, improving tractability
- Demonstrates controllable generation via compositional prompts, with potential for purely textual control in the future
- Shows diversity and motif recapitulation advantages over some baselines
- Cons
- Latent spaces in transformer-based models can be large and require regularization
- The approach relies on the quality of the underlying folding priors and the alignment between sequence and structure embeddings
- Real-world applicability depends on exposure to diverse function–organism prompts and robust decoding of complex structures
- Not all modalities or prompts may be equally well supported yet; broader multimodal extension remains an area of active development
Alternatives (brief comparisons)
| Approach | Modality | Strengths | Limitations |---|---|---|---| | PLAID | Sequence + structure | Co-generation with latent-diffusion; training on sequences; function/organism prompts | Requires a folding model as a backbone; quality depends on latent regularization |Traditional structure generators | Structure-only generation | Strong structural realism with folding priors | Lacks explicit sequence generation or multi-modal control; sequence compatibility not optimized |Sequence-only design models | Sequence generation only | Large sequence data coverage; fast exploration | No direct structural coordinates unless coupled with a separate predictor |
Pricing or License
Pricing or licensing details are not specified in the source.
References
- PLAID: Repurposing Protein Folding Models for Generation with Latent Diffusion — BAIR Blog, http://bair.berkeley.edu/blog/2025/04/08/plaid/
- PLAID and CHEAP preprints and codebases are referenced in the source; explicit URLs are not provided here.
More resources
Getting Started with NVIDIA Isaac for Healthcare Using the Telesurgery Workflow
A production-ready, modular telesurgery workflow from NVIDIA Isaac for Healthcare unifies simulation and clinical deployment across a low-latency, three-computer architecture. It covers video/sensor streaming, robot control, haptics, and simulation to support training and remote procedures.
NVFP4 Trains with Precision of 16-Bit and Speed and Efficiency of 4-Bit
NVFP4 is a 4-bit data format delivering FP16-level accuracy with the throughput and memory efficiency of 4-bit precision, extended to pretraining for large language models. This profile covers 12B-scale experiments, stability, and industry collaborations.
Inside NVIDIA Blackwell Ultra: The Chip Powering the AI Factory Era
An in‑depth profile of NVIDIA Blackwell Ultra, its dual‑die NV‑HBI design, NVFP4 precision, 288 GB HBM3e per GPU, and system‑level interconnects powering AI factories and large‑scale inference.
NVIDIA NeMo-RL Megatron-Core: Optimized Training Throughput
Overview of NeMo-RL v0.3 with Megatron-Core backend for post-training large models, detailing 6D/4D parallelism, GPU-optimized kernels, and simplified configuration to boost reinforcement learning throughput on models at scale.
Nemotron Nano 2 9B: Open Reasoning Model with 6x Throughput for Edge and Enterprise
Open Nemotron Nano 2 9B delivers leading accuracy and up to 6x throughput with a Hybrid Transformer–Mamba backbone and a configurable thinking budget, aimed at edge, PC and enterprise AI agents.
Accelerate ND-Parallel: A Guide to Efficient Multi-GPU Training
A concise resource on composing multiple parallelism strategies (DP, FSDP, TP, CP) with Accelerate and Axolotl to train large models across many GPUs, with guidance on configuration, use cases, and trade‑offs.





