Accelerate Protein Structure Inference Over 100x with NVIDIA RTX PRO 6000 Blackwell Server Edition
Sources: https://developer.nvidia.com/blog/accelerate-protein-structure-inference-over-100x-with-nvidia-rtx-pro-6000-blackwell-server-edition, https://developer.nvidia.com/blog/accelerate-protein-structure-inference-over-100x-with-nvidia-rtx-pro-6000-blackwell-server-edition/, NVIDIA Dev Blog
TL;DR
- The NVIDIA RTX PRO 6000 Blackwell Server Edition accelerates end-to-end protein structure inference on a single server using OpenFold, with accuracy unchanged relative to AlphaFold2.
- Benchmarks show up to 138x faster folding than AlphaFold2 and about 2.8x faster than ColabFold; alignment and inference speedups exceed hundreds of times versus CPU baselines.
- The platform provides 96 GB of high-bandwidth memory (1.6 TB/s), Multi-Instance GPU (MIG) capability that makes a single card act like four GPUs, and end-to-end, GPU-resident workflows for large MSAs and protein ensembles.
Context and background
Understanding protein structure is central to accelerating drug discovery, enzyme engineering, and advancing agricultural biotech. Since AlphaFold2 reshaped AI inference for protein structure, researchers have faced bottlenecks that limit speed and scale. CPU-bound multiple sequence alignment (MSA) generation and inefficient GPU inference have driven compute costs and extended project timelines. In this context, NVIDIA and its Digital Biology Research labs have focused on accelerating the full inference pipeline—from MSA generation through final structure prediction—without sacrificing accuracy. The result is a capability that enables large-scale protein analysis with OpenFold on powerful RTX PRO hardware, making proteome-scale folding accessible to labs, software platforms, and cloud providers alike. For context on the magnitude of the improvement, consider that optimized MMseqs2-GPU alignments can outperform CPU methods by orders of magnitude, and the combination of new hardware and software optimizations yields end-to-end speedups that redefine practical protein structure inference on a server scale. NVIDIA blog. Modern protein folding workloads often involve metagenomic-scale MSAs, iterative refinements, and ensemble calculations that can require hours of compute per target. Scaling these workloads across entire proteomes or drug-target libraries on CPU-based infrastructure is typically prohibitive. The GPU-based acceleration demonstrated with RTX PRO 6000 Blackwell addresses these bottlenecks by moving key components of the workflow onto a single, powerful GPU server, enabling rapid iteration and discovery in drug development, agriculture, and pandemic preparedness research. In benchmarks, MMseqs2-GPU achieved extraordinary gains versus CPU counterparts, underscoring the potential for GPU-centric pipelines to transform practical biology workloads.
What’s new
The latest NVIDIA RTX PRO 6000 Blackwell Server Edition introduces a combination of hardware and software innovations that push protein structure inference to new speeds:
- A high-performance GPU platform purpose-built for end-to-end inference with OpenFold, accelerated by new instructions and TensorRT optimizations.
- Efficient MMseqs2-GPU integration that dramatically speeds up MSA generation and pre-processing steps, enabling faster overall inference.
- Bespoke TensorRT optimizations targeting OpenFold, delivering significant speedups over baseline OpenFold inference on the same hardware.
- Validation across standard benchmarks, including 20 CASP14 protein targets, showing folding performance with equivalent TM-scores to AlphaFold2 while dramatically reducing wall time.
- High-bandwidth memory and MIG functionality: 96 GB of HBM with 1.6 TB/s bandwidth enables folding of large ensembles and MSAs entirely on the GPU, and MIG allows a single RTX PRO 6000 Blackwell to behave as four discrete GPUs, supporting multiple users or workflows on a single server without compromising speed or accuracy.
- Availability today: the RTX PRO 6000 Blackwell Server Edition is shipping with NVIDIA RTX PRO Servers from major system makers and in cloud instances from leading cloud providers.
Why it matters (impact for developers/enterprises)
For developers building software platforms for drug discovery, proteomics, or pandemic preparedness, this advancement translates into meaningful business and research benefits:
- Faster time to first predictions enables more rapid hypothesis testing, iterative design, and decision-making across research programs.
- Proteome-scale folding and large MSA handling become practical on a single server, reducing the need for sprawling CPU clusters and enabling smaller teams to perform large-scale analyses.
- The ability to run a full workflow on a GPU-resident server reduces data movement and accelerates iterative experimentation, which can shorten development cycles for new therapeutics, crop improvements, and biosurveillance tools.
- MIG enables multi-user collaboration on a shared server without sacrificing throughput, improving resource utilization in labs, cloud environments, and research centers.
- The combination of OpenFold, cuEquivariance, TensorRT, and MMseqs2-GPU on a single device sets a new baseline for end-to-end protein structure prediction speed, with practical implications for both research and product development. NVIDIA notes that these accelerations preserve the accuracy of predictions relative to AlphaFold2, making it feasible to adopt these workflows in production settings. NVIDIA blog.
Technical details or Implementation
The performance story rests on a combination of hardware capabilities and software optimizations that work together to deliver exceptional throughput:
- Hardware foundation: RTX PRO 6000 Blackwell Server Edition features 96 GB of high-bandwidth memory with 1.6 TB/s bandwidth, enabling the full workflow to remain GPU-resident, including large MSAs and protein ensembles. The architecture also supports Multi-Instance GPU (MIG) so a single GPU can be partitioned into four virtual GPUs.
- Software stack and optimizations: New instructions and TensorRT optimizations targeting OpenFold, together with MMseqs2-GPU acceleration, drive the major speedups. OpenFold benefits from bespoke TensorRT tuning, yielding a 2.3x improvement in inference speed over a baseline OpenFold setup.
- End-to-end speedups demonstrated on benchmarks: The NVIDIA Digital Biology Research lab validated OpenFold on RTX PRO 6000 Blackwell Server Edition with TensorRT. Overall, the folding workload on this platform achieved 138x faster performance than AlphaFold2 and about 2.8x faster than ColabFold, while preserving identical TM-scores. In a separate alignment benchmark, MMseqs2-GPU on a single L40S delivered ~177x faster alignments than CPU-based JackHMMER on a 128-core CPU, with up to 720x faster performance when distributed across eight L40S GPUs.
- Workflow integration: A complete example demonstrates deploying the OpenFold2 NIM on a local machine, constructing inference requests, and using a local endpoint to generate protein structure predictions. This enables folding on a single server at world-class speed, making proteome-scale analysis accessible to laboratories and software platforms alike.
- Availability and deployment: The RTX PRO 6000 Blackwell Server Edition is available today in NVIDIA RTX PRO Servers via global system makers and in cloud instances from leading cloud service providers. NVIDIA encourages potential partners to engage to realize protein folding at unprecedented speed and scale. The acknowledgments include researchers from NVIDIA, University of Oxford, and Seoul National University.
Key takeaways
- End-to-end protein structure inference can now run on a single server with RTX PRO 6000 Blackwell, delivering world-class speed without accuracy loss relative to AlphaFold2.
- Speedups span the entire pipeline: MMseqs2-GPU accelerates MSA generation, TensorRT optimizations accelerate OpenFold, and the combined result far outpaces CPU-based workflows.
- Benchmarks show fold performance up to 138x faster than AlphaFold2 and 2.8x faster than ColabFold, with 177x faster sequence alignments on a single L40S and up to 720x with multiple GPUs.
- Hardware features such as 96 GB HBM and MIG enable large-scale, GPU-resident workflows and multi-user sharing on a single server.
- Availability is immediate today through NVIDIA RTX PRO Servers and cloud service providers, enabling labs and platforms to adopt proteome-scale folding workflows.
FAQ
-
What is the RTX PRO 6000 Blackwell Server Edition capable of powering?
It enables end-to-end protein structure inference with OpenFold on a single server, leveraging MMseqs2-GPU, TensorRT, and other accelerations to deliver substantial speedups over AlphaFold2 and ColabFold while preserving accuracy.
-
How fast is the new solution compared with established baselines?
It folds content up to 138x faster than AlphaFold2 and about 2.8x faster than ColabFold for the OpenFold workflow; MMseqs2-GPU aligns sequences ~177x faster on a single L40S versus CPU JackHMMER, with up to 720x speedups when scaling to eight GPUs.
-
What hardware features support these gains?
The platform provides 96 GB of high-bandwidth memory (1.6 TB/s), MIG to partition a single GPU into four virtual GPUs, and a GPU-focused stack including cuEquivariance, TensorRT, and MMseqs2-GPU.
-
Is this available now?
Yes. The RTX PRO 6000 Blackwell Server Edition is available today in NVIDIA RTX PRO Servers from global system makers and in cloud instances from leading cloud service providers.
-
Where can I learn more or start a deployment?
See the NVIDIA blog for detailed benchmarks, results, and deployment guidance, and connect with NVIDIA partners to plan a setup tailored to laboratory or enterprise needs. [NVIDIA blog](https://developer.nvidia.com/blog/accelerate-protein-structure-inference-over-100x-with-nvidia-rtx-pro-6000-blackwell-server-edition/).
References
More news
NVIDIA HGX B200 Reduces Embodied Carbon Emissions Intensity
NVIDIA HGX B200 lowers embodied carbon intensity by 24% vs. HGX H100, while delivering higher AI performance and energy efficiency. This article reviews the PCF-backed improvements, new hardware features, and implications for developers and enterprises.
Predict Extreme Weather in Minutes Without a Supercomputer: Huge Ensembles (HENS)
NVIDIA and Berkeley Lab unveil Huge Ensembles (HENS), an open-source AI tool that forecasts low-likelihood, high-impact weather events using 27,000 years of data, with ready-to-run options.
Scaleway Joins Hugging Face Inference Providers for Serverless, Low-Latency Inference
Scaleway is now a supported Inference Provider on the Hugging Face Hub, enabling serverless inference directly on model pages with JS and Python SDKs. Access popular open-weight models and enjoy scalable, low-latency AI workflows.
How to Reduce KV Cache Bottlenecks with NVIDIA Dynamo
NVIDIA Dynamo offloads KV Cache from GPU memory to cost-efficient storage, enabling longer context windows, higher concurrency, and lower inference costs for large-scale LLMs and generative AI workloads.
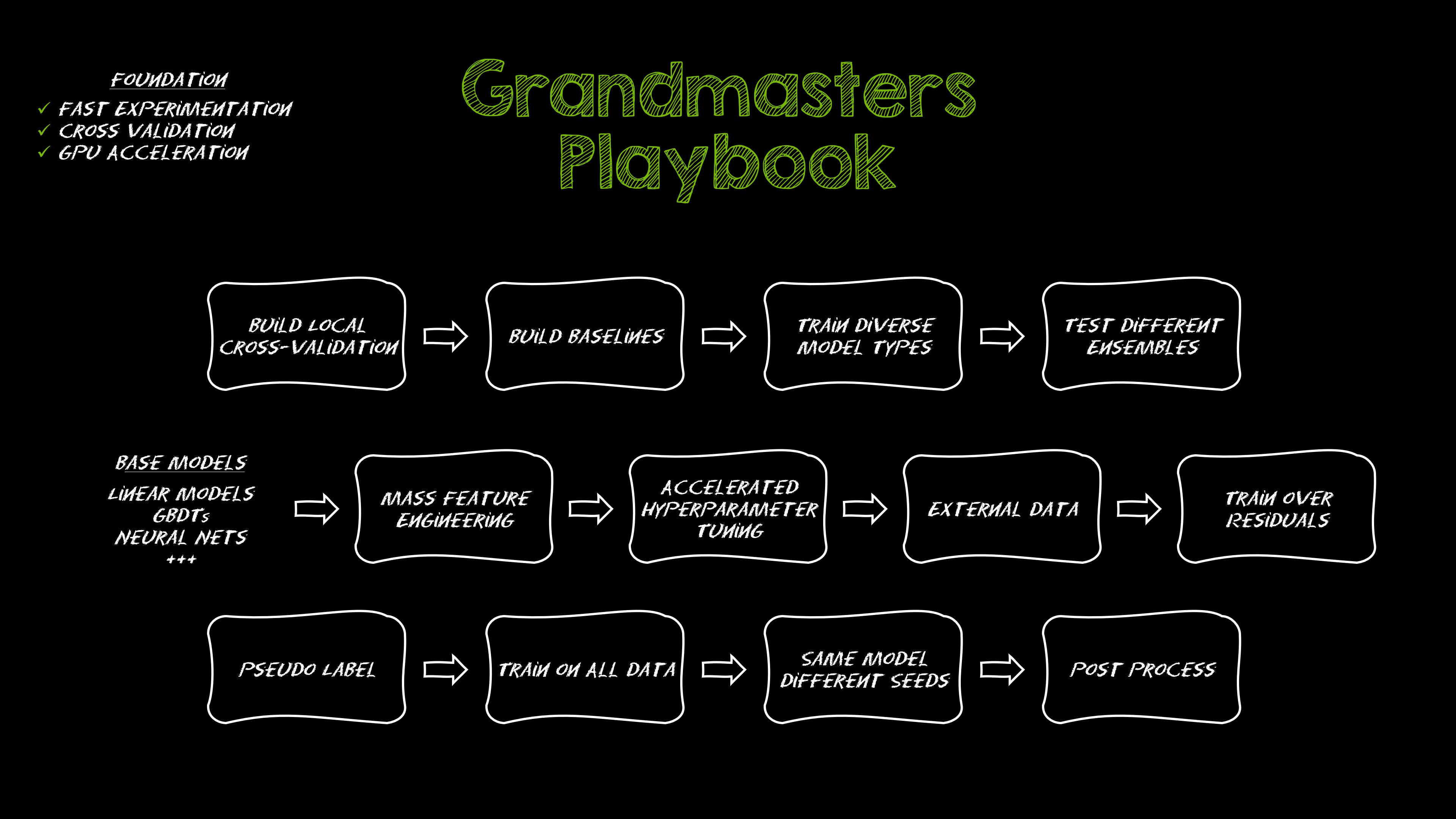
Kaggle Grandmasters Playbook: 7 Battle-Tested Techniques for Tabular Data Modeling
A detailed look at seven battle-tested techniques used by Kaggle Grandmasters to solve large tabular datasets fast with GPU acceleration, from diversified baselines to advanced ensembling and pseudo-labeling.
Microsoft to turn Foxconn site into Fairwater AI data center, touted as world's most powerful
Microsoft unveils plans for a 1.2 million-square-foot Fairwater AI data center in Wisconsin, housing hundreds of thousands of Nvidia GB200 GPUs. The project promises unprecedented AI training power with a closed-loop cooling system and a cost of $3.3 billion.