Parquet Content-Defined Chunking with Xet: Faster uploads and smarter deduplication
Sources: https://huggingface.co/blog/parquet-cdc, Hugging Face Blog
TL;DR
- Parquet Content-Defined Chunking (CDC) is now available in PyArrow and Pandas, enabling efficient deduplication on Hugging Face’s Xet storage layer.
- CDC deduplicates data across repositories, reducing both upload and download data transfer and overall storage costs.
- Enable the feature by passing use_content_defined_chunking=True to the Parquet writer (PyArrow) and is supported in Pandas as well.
- The approach leverages content-based chunking before serialization and compression, improving efficiency for Parquet workloads across the Hub.
- The OpenOrca demonstration shows substantial reductions in transferred bytes and faster operations when CDC is enabled.
Context and background
Parquet is a widely used columnar storage format. Hugging Face hosts nearly 21 petabytes of datasets, with Parquet files accounting for over 4 petabytes of that storage. To address the scale of this data, Hugging Face introduced a new storage layer called Xet, which uses content-defined chunking to deduplicate chunks of data. This approach reduces storage costs and speeds up uploads and downloads, particularly when multiple workstreams share similar or evolving data. Content-defined chunking (CDC) sits alongside Xet’s deduplication capabilities. Parquet’s layout—organized by columns and column chunks (data pages) with compression—can yield different byte-level representations for small changes. The CDC feature mitigates this by chunking data pages based on their content, aligning the deduplication mechanism with the logical data values rather than the raw serialized bytes. This alignment helps Xet more effectively recognize when data remains the same despite minor edits. The article also notes that since PyArrow 21.0.0, Hugging Face URIs (hf://) can be used directly in PyArrow for reading and writing Parquet (and other formats) to the Hub, broadening programmatic access for data engineers and data scientists alike. The combination of Parquet CDC and the Xet storage layer enables cross-repository and cross-file deduplication, which is crucial for collaborative workflows and large-scale dataset management.
What’s new
The Parquet CDC feature is now available in both PyArrow and Pandas, allowing users to write Parquet files with content-defined chunking enabled. The key operational change is the ability to write with use_content_defined_chunking=True, which ensures that columns are chunked into data pages according to their content, before serialization or compression. This mirrors how the Xet storage layer deduplicates data, but applies the deduplication logic to the logical values of the columns. Demonstrations on a manageable OpenOrca subset show that the original write and subsequent reads can be performed using Hugging Face URIs (hf://) in PyArrow. When the same file is uploaded again or to a different repository, deduplication can recognize identical content and upload only the changed portions, resulting in near-instantaneous operations because no unnecessary data is transferred. This cross-repository deduplication is a hallmark of the Xet storage layer and is a core advantage highlighted in the CDC-enabled workflow. A closer look at the behavior with CDC reveals several practical benefits:
- Only new columns and updated footer metadata are uploaded when columns are added.
- Removing columns similarly affects only the footer, with unchanged data remaining on the storage layer.
- Changing a column type (e.g., int64 to int32) transfers only the new column data and updated metadata.
- Appending new rows transfers only the new rows, with multiple red regions appearing in heatmaps due to the Parquet data-page layout, yet still avoiding a full re-upload of existing data.
- Writing Parquet files across multiple, differently bounded files (sharding) with CDC maintains dedup across files and boundaries, with the overall uploaded size scarcely larger than the original dataset. These observations are demonstrated in the blog post and illustrated with heatmaps that visualize deduplication progress and transferred data.
Why it matters (impact for developers/enterprises)
For teams working with large Parquet datasets on Hugging Face, CDC implemented via the Xet storage layer offers meaningful advantages:
- Reduced data transfer: By deduplicating at the content level, only changed data pages and footer metadata are exchanged during uploads and downloads.
- Lower storage costs: Cross-repository deduplication helps avoid storing multiple copies of identical data, especially as datasets evolve or are reused across projects.
- Faster collaboration: Efficient data sharing and dedup across repositories accelerates workflows that involve sharing and recombining datasets.
- Read/write performance benefits: The combination of Parquet CDC with the Xet layer optimizes common Parquet operations, including reads and writes, and can scale with dataset size and complexity.
- Broad applicability: The Xet storage layer is format-agnostic, while Parquet CDC targets the specific data-page level chunking of Parquet to maximize deduplication effectiveness. For practitioners, these benefits translate into shorter iteration cycles, lower cloud egress or storage costs, and more responsive data pipelines as datasets grow and evolve. The integration also aligns with PyArrow and Pandas ecosystems, making CDC accessible to a wide range of data engineers and data scientists. Hugging Face Blog
Technical details or Implementation (how to implement)
- Enabling CDC: Parquet CDC can be enabled by passing use_content_defined_chunking=True to the write_parquet function in PyArrow. Pandas also supports the new feature, enabling CDC-enabled Parquet writes from Python with familiar APIs.
- URIs and access: PyArrow supports using Hugging Face URIs (hf://) to read and write Parquet files directly to the Hub since PyArrow 21.0.0, simplifying integration into data workflows.
- Data-page level chunking: CDC operates at the Parquet data page (column chunk) level, chunking columns into pages based on content rather than raw bytes, aligning with the Xet deduplication logic.
- Row-group considerations: Parquet writers use fixed row-group sizes by default (e.g., PyArrow defaults to 1 Mi rows). Dataset writers may reduce the row-group size to improve random access performance or reduce memory usage; changing row-group size can shift data between pages and affect dedup results similarly to insertions/deletions.
- Cross-file deduplication: Parquet CDC combined with the Xet storage layer can efficiently deduplicate data across multiple files, even when data is split across different boundaries (shards).
- Practical scenarios: The CDC approach shows that adding new columns, removing columns, or changing column types can minimize transferred bytes and metadata, while appending rows transfers only the new data—though the exact dedup ratio depends on the distribution of changes across data pages. | Scenario | Deduplication behavior with Parquet CDC + Xet |--- |--- |New columns added | Only the new columns and updated footer metadata are uploaded; original data remains on storage |Removing columns | Only footer metadata changes; other columns remain in storage and are not transferred again |Changing a column type (e.g., int64 to int32) | Only the new column data and updated metadata are uploaded |Appending new rows | Only the new rows are transferred; multiple red regions may appear due to per-column data-page layout |Cross-file and sharding | Deduplication works across files and boundaries; overall upload size remains nearly the original size |
- Download considerations: The same CDC benefits apply to downloads via huggingface_hub.hf_hub_download() and datasets.load_dataset() functions, reinforcing end-to-end efficiency for data access.
- Limitations and caveats: The effectiveness of deduplication with CDC depends on the reader/writer constraints and the distribution of changes across the dataset. In cases where most data pages are affected by updates, the deduplication ratio may decline.
Key takeaways
- Parquet CDC is now available in PyArrow and Pandas and works with Hugging Face’s Xet storage layer to deduplicate data at the column data-page level before compression.
- Enabling CDC requires a simple flag (use_content_defined_chunking=True) during Parquet writes, with cross-repository deduplication benefiting collaborative workflows.
- The approach reduces both data transfer and storage costs and improves performance for Parquet operations on the Hub, including reads and writes across multiple files and repositories.
- CDC’s effectiveness depends on the nature of changes; most column-level changes can be handled efficiently, while heavy modifications may reduce dedup efficiency. Practical demonstrations indicate substantial transfer reductions when CDC is enabled.
- The Xet storage layer is format-agnostic, and CDC complements this capability by applying content-aware chunking to Parquet data before serialization and compression, enabling more effective data sharing and collaboration. Hugging Face Blog
FAQ
-
What is Parquet Content-Defined Chunking (CDC)?
CDC is a PyArrow feature (also supported in Pandas) that chunks Parquet data pages based on content, enabling dedup across the Xet storage layer by operating on logical column values before serialization or compression.
-
How do I enable Parquet CDC?
In PyArrow, pass use_content_defined_chunking=True to the write_parquet function; Pandas also supports the feature for Parquet writes.
-
Does CDC affect downloads as well as uploads?
Yes. The same deduplication benefits apply to downloads via hf_hub_download() and datasets.load_dataset(), improving transfer efficiency.
-
How does row-group size influence CDC performance?
Parquet writers use fixed-sized row-groups by default (e.g., 1 Mi rows in PyArrow). Depending on the reader/writer constraints, smaller or larger row-groups may be beneficial for access patterns, but changing row-groups can shift data pages and impact deduplication similarly to edits in the dataset.
-
Can CDC deduplicate data across multiple files?
Yes. Parquet CDC combined with Xet can efficiently deduplicate data across multiple files, even when data is split across different boundaries, with overall upload size remaining close to the original. [Hugging Face Blog](https://huggingface.co/blog/parquet-cdc)
References
More news
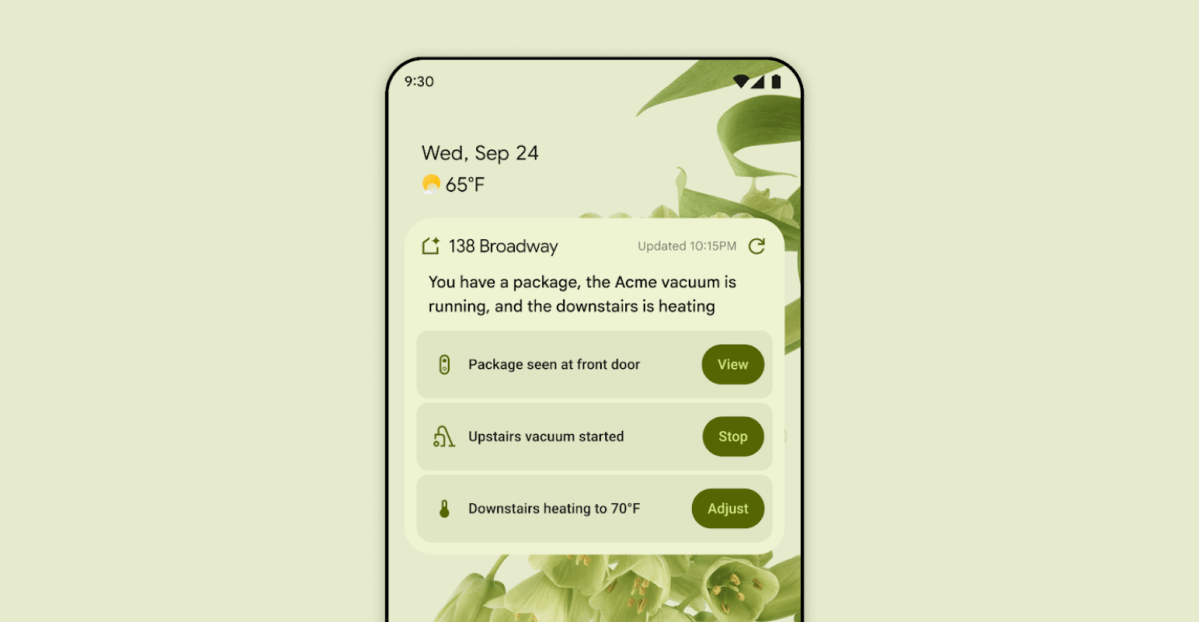
First look at the Google Home app powered by Gemini
The Verge reports Google is updating the Google Home app to bring Gemini features, including an Ask Home search bar, a redesigned UI, and Gemini-driven controls for the home.
Shadow Leak shows how ChatGPT agents can exfiltrate Gmail data via prompt injection
Security researchers demonstrated a prompt-injection attack called Shadow Leak that leveraged ChatGPT’s Deep Research to covertly extract data from a Gmail inbox. OpenAI patched the flaw; the case highlights risks of agentic AI.
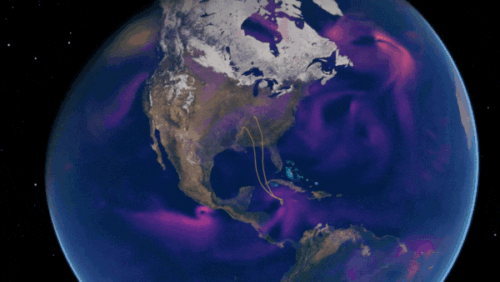
Predict Extreme Weather in Minutes Without a Supercomputer: Huge Ensembles (HENS)
NVIDIA and Berkeley Lab unveil Huge Ensembles (HENS), an open-source AI tool that forecasts low-likelihood, high-impact weather events using 27,000 years of data, with ready-to-run options.
Scaleway Joins Hugging Face Inference Providers for Serverless, Low-Latency Inference
Scaleway is now a supported Inference Provider on the Hugging Face Hub, enabling serverless inference directly on model pages with JS and Python SDKs. Access popular open-weight models and enjoy scalable, low-latency AI workflows.
Google expands Gemini in Chrome with cross-platform rollout and no membership fee
Gemini AI in Chrome gains access to tabs, history, and Google properties, rolling out to Mac and Windows in the US without a fee, and enabling task automation and Workspace integrations.
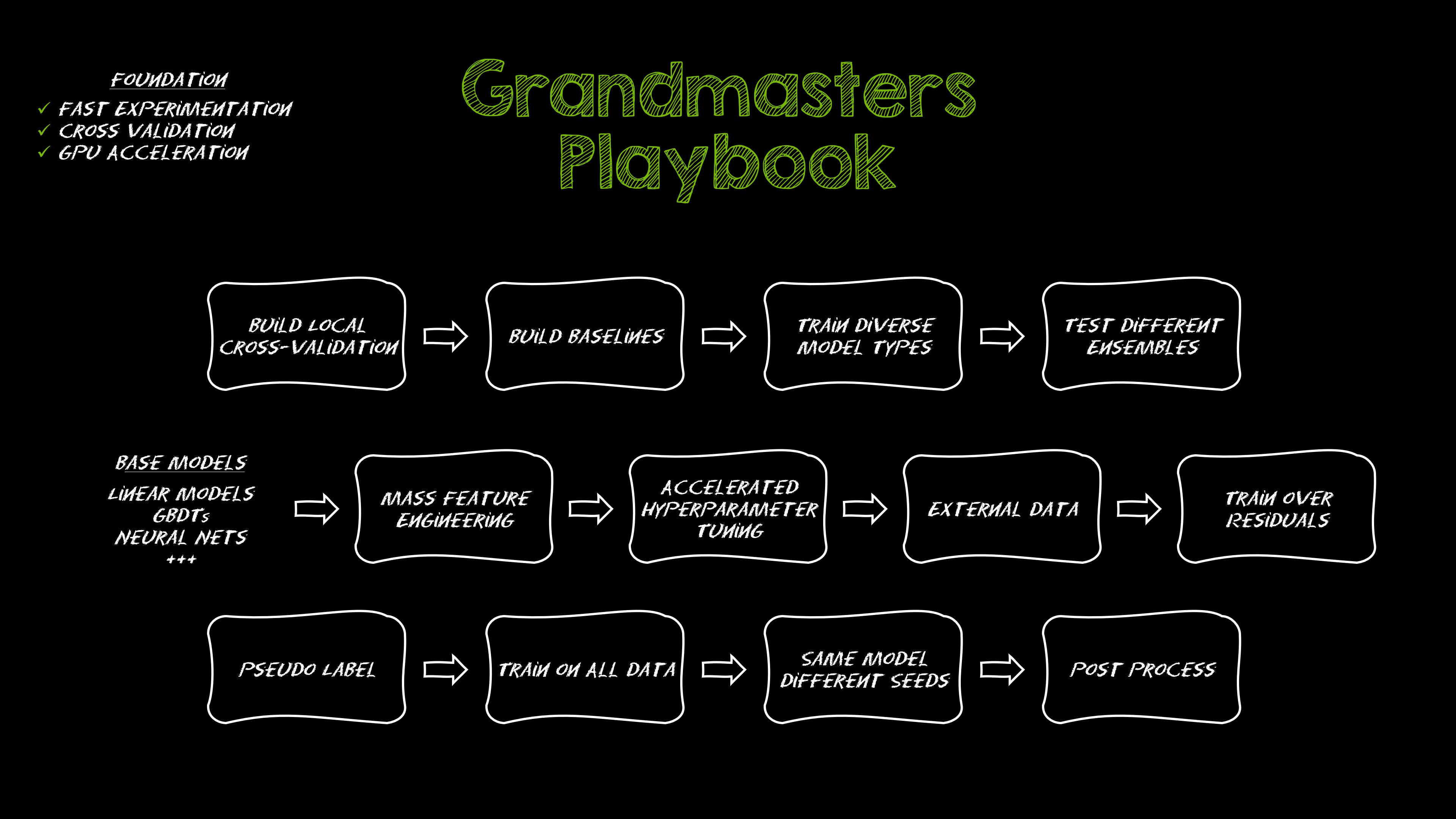
Kaggle Grandmasters Playbook: 7 Battle-Tested Techniques for Tabular Data Modeling
A detailed look at seven battle-tested techniques used by Kaggle Grandmasters to solve large tabular datasets fast with GPU acceleration, from diversified baselines to advanced ensembling and pseudo-labeling.